Clustering#
Import des outils / jeu de données#
1import matplotlib.pyplot as plt
2import numpy as np
3import pandas as pd
4import prince
5import seaborn as sns
6from sklearn.cluster import (
7 DBSCAN,
8 OPTICS,
9 AffinityPropagation,
10 AgglomerativeClustering,
11 KMeans,
12 MeanShift,
13)
14from sklearn.compose import ColumnTransformer
15from sklearn.metrics import (
16 calinski_harabasz_score,
17 davies_bouldin_score,
18 silhouette_score,
19)
20from sklearn.mixture import GaussianMixture
21from sklearn.preprocessing import RobustScaler, StandardScaler
22
23from src.clustering import initiate_cluster_models
24from src.config import data_folder, seed
25from src.constants import var_categoriques, var_numeriques
26from src.utils import init_notebook
1init_notebook()
1df = pd.read_csv(
2 f"{data_folder}/data-cleaned-feature-engineering.csv",
3 sep=",",
4 index_col="ID",
5 parse_dates=True,
6)
1composantes_acp = pd.read_csv(f"{data_folder}/composantes_acp.csv", index_col="ID")
2composantes_acm = pd.read_csv(f"{data_folder}/composantes_acm.csv", index_col="ID")
Variables globales#
1var_categoriques_extra = ["NbAcceptedCampaigns", "HasAcceptedCampaigns", "NbChildren"]
2
3var_categoriques_fe = var_categoriques + var_categoriques_extra
Clustering#
Préparation des données#
Nous commencer par fusionner les variables quantitatives et les coordonnées des individus dans l’ACM.
1X_clust = pd.concat((df[var_numeriques], composantes_acm), axis=1)
1X_clust.head()
| Year_Birth | Income | Recency | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumDealsPurchases | ... | ACM4 | ACM5 | ACM6 | ACM7 | ACM8 | ACM9 | ACM10 | ACM11 | ACM12 | ACM13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | |||||||||||||||||||||
| 5524 | 1957 | 58138.0 | 58 | 635 | 88 | 546 | 172 | 88 | 88 | 3 | ... | -0.909429 | 0.439397 | 0.662880 | -0.292805 | -0.039068 | 0.196400 | -0.843529 | -0.898797 | 0.174169 | 0.960631 |
| 2174 | 1954 | 46344.0 | 38 | 11 | 1 | 6 | 2 | 1 | 6 | 2 | ... | -0.402198 | 0.129620 | 0.171945 | -0.143049 | -0.116974 | 0.105742 | 0.259841 | 0.262817 | -0.093681 | 0.033373 |
| 4141 | 1965 | 71613.0 | 26 | 426 | 49 | 127 | 111 | 21 | 42 | 1 | ... | -0.423124 | -0.465975 | 0.553202 | 0.541437 | -1.880626 | 0.028891 | -0.374510 | -0.919603 | 0.287754 | -0.350058 |
| 6182 | 1984 | 26646.0 | 26 | 11 | 4 | 20 | 10 | 3 | 5 | 2 | ... | -0.275507 | -0.245603 | 0.246638 | 0.239619 | -0.942750 | -0.002593 | -0.333331 | 0.052414 | -0.412546 | -0.177088 |
| 5324 | 1981 | 58293.0 | 94 | 173 | 43 | 118 | 46 | 27 | 15 | 5 | ... | 0.554819 | 0.142063 | -0.591854 | -0.680550 | 0.175540 | -0.518129 | -0.348914 | 0.007669 | -0.491990 | -0.319610 |
5 rows × 27 columns
1preprocessor = ColumnTransformer(
2 remainder="passthrough",
3 transformers=[
4 ("scaler", RobustScaler(), var_numeriques),
5 ],
6)
1scaler = RobustScaler()
2df_apres_scale = pd.DataFrame(
3 preprocessor.fit_transform(X_clust),
4 columns=X_clust.columns,
5 index=df.index,
6)
1df_apres_scale.head()
| Year_Birth | Income | Recency | MntWines | MntFruits | MntMeatProducts | MntFishProducts | MntSweetProducts | MntGoldProds | NumDealsPurchases | ... | ACM4 | ACM5 | ACM6 | ACM7 | ACM8 | ACM9 | ACM10 | ACM11 | ACM12 | ACM13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | |||||||||||||||||||||
| 5524 | -0.722222 | 0.187662 | 0.18 | 0.957469 | 2.50000 | 2.238318 | 3.386243 | 2.424242 | 1.361702 | 0.5 | ... | -0.909429 | 0.439397 | 0.662880 | -0.292805 | -0.039068 | 0.196400 | -0.843529 | -0.898797 | 0.174169 | 0.960631 |
| 2174 | -0.888889 | -0.175314 | -0.22 | -0.337137 | -0.21875 | -0.285047 | -0.211640 | -0.212121 | -0.382979 | 0.0 | ... | -0.402198 | 0.129620 | 0.171945 | -0.143049 | -0.116974 | 0.105742 | 0.259841 | 0.262817 | -0.093681 | 0.033373 |
| 4141 | -0.277778 | 0.602373 | -0.46 | 0.523859 | 1.28125 | 0.280374 | 2.095238 | 0.393939 | 0.382979 | -0.5 | ... | -0.423124 | -0.465975 | 0.553202 | 0.541437 | -1.880626 | 0.028891 | -0.374510 | -0.919603 | 0.287754 | -0.350058 |
| 6182 | 0.777778 | -0.781546 | -0.46 | -0.337137 | -0.12500 | -0.219626 | -0.042328 | -0.151515 | -0.404255 | 0.0 | ... | -0.275507 | -0.245603 | 0.246638 | 0.239619 | -0.942750 | -0.002593 | -0.333331 | 0.052414 | -0.412546 | -0.177088 |
| 5324 | 0.611111 | 0.192433 | 0.90 | -0.001037 | 1.09375 | 0.238318 | 0.719577 | 0.575758 | -0.191489 | 1.5 | ... | 0.554819 | 0.142063 | -0.591854 | -0.680550 | 0.175540 | -0.518129 | -0.348914 | 0.007669 | -0.491990 | -0.319610 |
5 rows × 27 columns
1df_avec_clusters = df_apres_scale.copy()
Différents algorithmes de clustering#
Nous choisissons de tester 2 types de modèles de clustering :
les modèles à choix du nombre de clusters
les modèles qui décident du nombre de clusters
Cela nous permettra de comparer le nombre de clusters donné par les seconds algorithmes.
Pour les modèles pour lesquels il faut choisir le nombre de clusters, nous décidons de tester des clusters de taille 2 à 5 (inclus), car un trop grand nombre de clusters serait plus difficile à interpréter pour l’équipe marketing dans un premier temps.
Tableau. Méthodologie de clustering
|:—————————-|:—-|
| Algorithmes | Avec choix du nombre de clusters (entre 2 et 5)
Sans choix du nombre de clusters |
| Critères de sélection | Répartition des clusters
Métriques de clusters
Sélection manuelle des clusters via leur affichage |
| Métriques | Score Silhouette (entre -1 et 1, proche de 1 = meilleurs clusters)
Calinski-Harabasz (entre 0 et $+\infty$ plus grand = meilleure séparation)
Davies-Bouldin (entre 0 et $+\infty$, proche de 0 = meilleurs clusters) |
| Affichage des clusters | Sur les axes d’ACP 1-4
Sur les axes d’ACM 1-4
En fonction des variables quantitatives
En fonction des variables qualitatives |
Tableau. Algorithmes de clustering testés
Choix du nombre de clusters |
Algorithmes |
|---|---|
Avec |
KMeans |
Sans |
OPTICS |
1NB_CLUSTER_MIN = 2
2NB_CLUSTER_MAX = 6 ## non inclus
1model_clusters = initiate_cluster_models(
2 NB_CLUSTER_MIN,
3 NB_CLUSTER_MAX,
4 seed,
5)
1a = GaussianMixture()
1isinstance(a, GaussianMixture)
True
1cluster_metrics = []
2
3for (model_name, model) in model_clusters.items():
4 if isinstance(model, GaussianMixture): ## cas particulier du mélange gaussien
5 df_avec_clusters[model_name] = model.fit_predict(df_apres_scale)
6 else:
7 model.fit(df_apres_scale)
8 df_avec_clusters[model_name] = model.labels_
9
10 df_avec_clusters[model_name] = pd.Categorical(
11 df_avec_clusters[model_name].astype(str)
12 )
13
14 nb_clusters = df_avec_clusters[model_name].nunique()
15
16 repartition = list(
17 df_avec_clusters[model_name].value_counts(normalize=True).round(2).astype(str)
18 ) ## todo: enlever astype(str) si ça sert à rien (tester)
19
20 cluster_metrics.append(
21 [
22 model_name,
23 nb_clusters,
24 " | ".join(repartition),
25 silhouette_score(
26 df_apres_scale, df_avec_clusters[model_name], random_state=seed
27 ), ## proche de 1 = mieux
28 calinski_harabasz_score(
29 df_apres_scale,
30 df_avec_clusters[model_name],
31 ), ## plus élevé, mieux c'est
32 davies_bouldin_score(
33 df_apres_scale, df_avec_clusters[model_name]
34 ), ## proche de 0 = mieux
35 ]
36 )
/home/runner/.cache/pypoetry/virtualenvs/customer-base-analysis-F-W2gxNr-py3.10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1416: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=10)
/home/runner/.cache/pypoetry/virtualenvs/customer-base-analysis-F-W2gxNr-py3.10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1416: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=10)
/home/runner/.cache/pypoetry/virtualenvs/customer-base-analysis-F-W2gxNr-py3.10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1416: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=10)
/home/runner/.cache/pypoetry/virtualenvs/customer-base-analysis-F-W2gxNr-py3.10/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py:1416: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
super()._check_params_vs_input(X, default_n_init=10)
/home/runner/.cache/pypoetry/virtualenvs/customer-base-analysis-F-W2gxNr-py3.10/lib/python3.10/site-packages/sklearn/cluster/_affinity_propagation.py:142: ConvergenceWarning: Affinity propagation did not converge, this model may return degenerate cluster centers and labels.
warnings.warn(
1pd.DataFrame(
2 cluster_metrics,
3 columns=[
4 "Algorithme de clustering",
5 "Nombre de clusters",
6 "Répartition",
7 "Silhouette",
8 "Calinski-Harabasz",
9 "Davies-Bouldin",
10 ],
11)
| Algorithme de clustering | Nombre de clusters | Répartition | Silhouette | Calinski-Harabasz | Davies-Bouldin | |
|---|---|---|---|---|---|---|
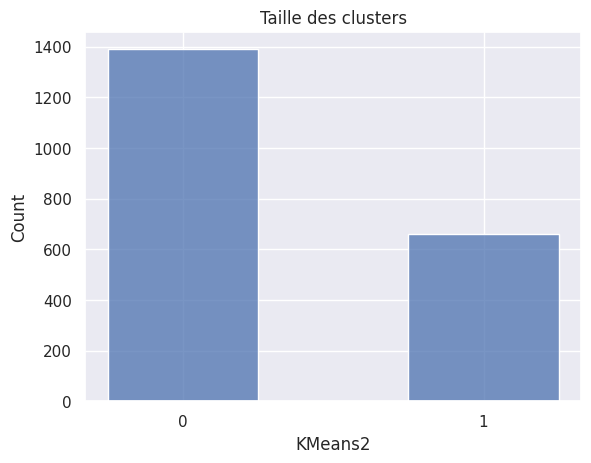
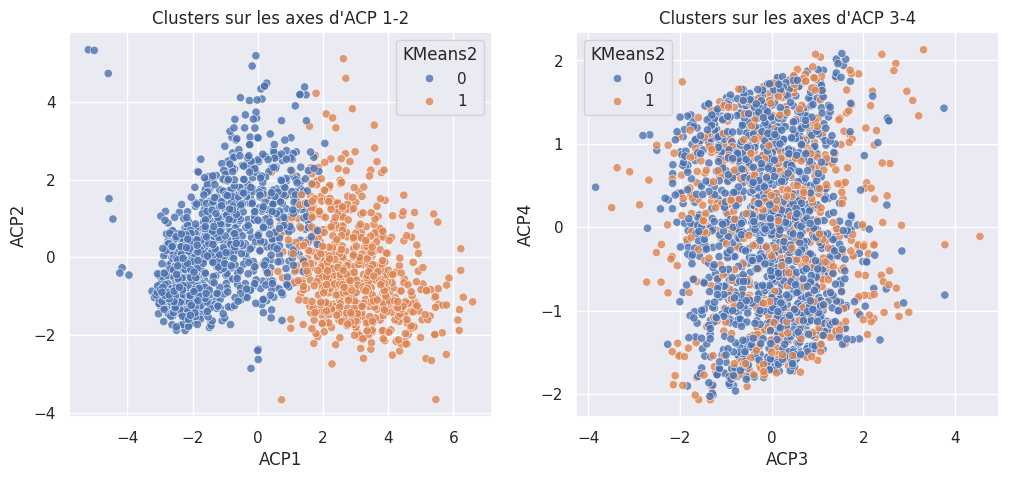
| 0 | KMeans2 | 2 | 0.68 | 0.32 | 0.318482 | 754.878090 | 1.547400 |
| 1 | KMeans3 | 3 | 0.46 | 0.29 | 0.25 | 0.167516 | 513.955410 | 1.982583 |
| 2 | KMeans4 | 4 | 0.45 | 0.25 | 0.19 | 0.12 | 0.157546 | 402.274688 | 2.334645 |
| 3 | KMeans5 | 5 | 0.44 | 0.23 | 0.19 | 0.12 | 0.02 | 0.170982 | 340.753601 | 2.058930 |
| 4 | GMM2 | 2 | 0.52 | 0.48 | 0.207188 | 542.083968 | 1.740252 |
| 5 | GMM3 | 3 | 0.52 | 0.31 | 0.18 | 0.062900 | 218.097499 | 3.099757 |
| 6 | GMM4 | 4 | 0.36 | 0.3 | 0.27 | 0.06 | 0.087929 | 268.462211 | 4.010402 |
| 7 | GMM5 | 5 | 0.43 | 0.34 | 0.13 | 0.07 | 0.02 | 0.129101 | 205.782852 | 3.352396 |
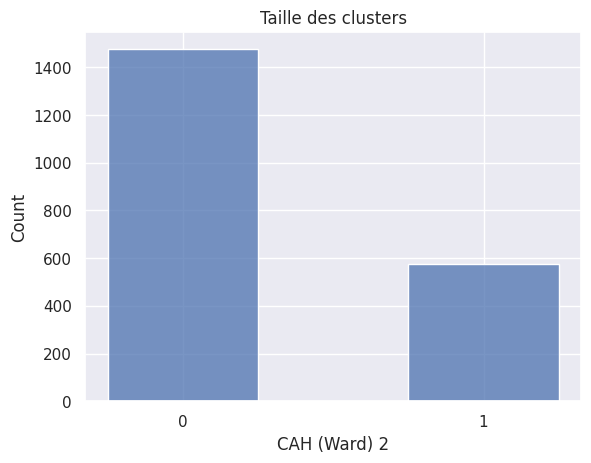
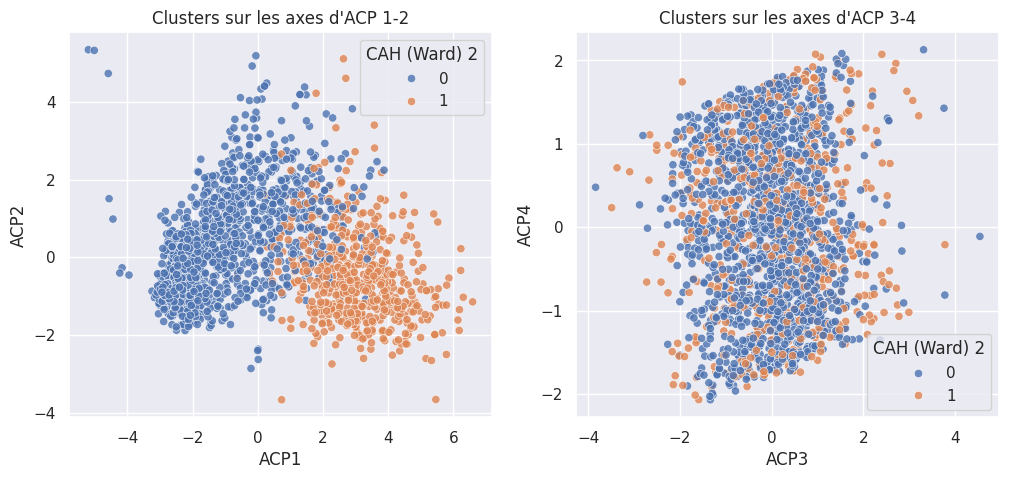
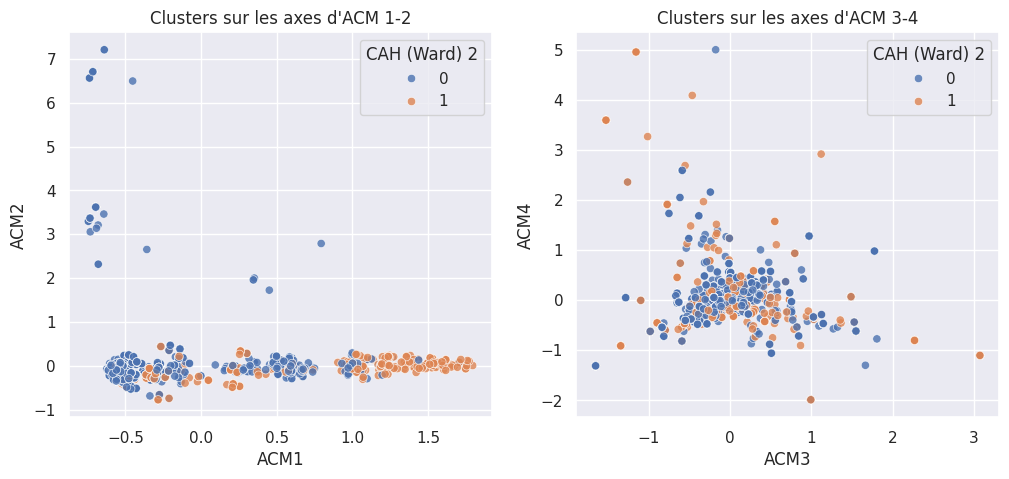
| 8 | CAH (Ward) 2 | 2 | 0.72 | 0.28 | 0.314124 | 657.064212 | 1.618113 |
| 9 | CAH (Ward) 3 | 3 | 0.44 | 0.28 | 0.28 | 0.138385 | 456.284028 | 2.071393 |
| 10 | CAH (Ward) 4 | 4 | 0.41 | 0.28 | 0.28 | 0.02 | 0.147383 | 348.454297 | 1.794500 |
| 11 | CAH (Ward) 5 | 5 | 0.41 | 0.28 | 0.22 | 0.06 | 0.02 | 0.142491 | 292.850286 | 2.186182 |
| 12 | CAH (average linkage) 2 | 2 | 1.0 | 0.0 | 0.552208 | 29.958308 | 0.928812 |
| 13 | CAH (average linkage) 3 | 3 | 1.0 | 0.0 | 0.0 | 0.523717 | 18.536309 | 0.735997 |
| 14 | CAH (average linkage) 4 | 4 | 1.0 | 0.0 | 0.0 | 0.0 | 0.478857 | 14.305294 | 0.755391 |
| 15 | CAH (average linkage) 5 | 5 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.458367 | 14.762254 | 0.896412 |
| 16 | CAH (single linkage) 2 | 2 | 1.0 | 0.0 | 0.587091 | 26.332043 | 0.549969 |
| 17 | CAH (single linkage) 3 | 3 | 1.0 | 0.0 | 0.0 | 0.568624 | 17.823639 | 0.468717 |
| 18 | CAH (single linkage) 4 | 4 | 1.0 | 0.0 | 0.0 | 0.0 | 0.540606 | 14.538207 | 0.434358 |
| 19 | CAH (single linkage) 5 | 5 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.518696 | 12.704006 | 0.417355 |
| 20 | CAH (complete linkage) 2 | 2 | 1.0 | 0.0 | 0.552208 | 29.958308 | 0.928812 |
| 21 | CAH (complete linkage) 3 | 3 | 1.0 | 0.0 | 0.0 | 0.540075 | 21.966639 | 0.879092 |
| 22 | CAH (complete linkage) 4 | 4 | 0.72 | 0.27 | 0.0 | 0.0 | 0.317609 | 248.338621 | 1.259276 |
| 23 | CAH (complete linkage) 5 | 5 | 0.71 | 0.27 | 0.01 | 0.0 | 0.0 | 0.290965 | 202.334927 | 1.242878 |
| 24 | OPTICS | 28 | 0.89 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01... | -0.295888 | 7.766509 | 1.447745 |
| 25 | MeanShift | 22 | 0.84 | 0.04 | 0.02 | 0.02 | 0.02 | 0.02 | 0.01... | 0.197199 | 42.398326 | 1.371609 |
| 26 | AffinityPropagation | 142 | 0.04 | 0.03 | 0.03 | 0.02 | 0.02 | 0.02 | 0.02... | 0.098856 | 40.876485 | 1.657521 |
Clusters sélectionnés :
KMeans 2
GMM 2
CAH (Ward 2)
Nous avons aussi étudié certains clusters avec 3 groupes, qui nous ont permis d’identifier certains individus, mais qui ne sont pas aussi intéressants et utilisables que les clusters avec 2 groupes.
Visualisation#
1def affiche_taille_clusters(nom_cluster):
2 plt.title("Taille des clusters")
3 sns.histplot(df_avec_clusters[nom_cluster], shrink=0.5)
4
5 plt.show()
1def affiche_clusters_acp(nom_cluster):
2 _, ax = plt.subplots(1, 2, figsize=(12, 5))
3
4 ax[0].set_title("Clusters sur les axes d'ACP 1-2")
5 ax[1].set_title("Clusters sur les axes d'ACP 3-4")
6
7 sns.scatterplot(
8 composantes_acp,
9 x="ACP1",
10 y="ACP2",
11 hue=df_avec_clusters[nom_cluster],
12 alpha=0.8,
13 ax=ax[0],
14 )
15 sns.scatterplot(
16 composantes_acp,
17 x="ACP3",
18 y="ACP4",
19 hue=df_avec_clusters[nom_cluster],
20 alpha=0.8,
21 ax=ax[1],
22 )
23
24 plt.show()
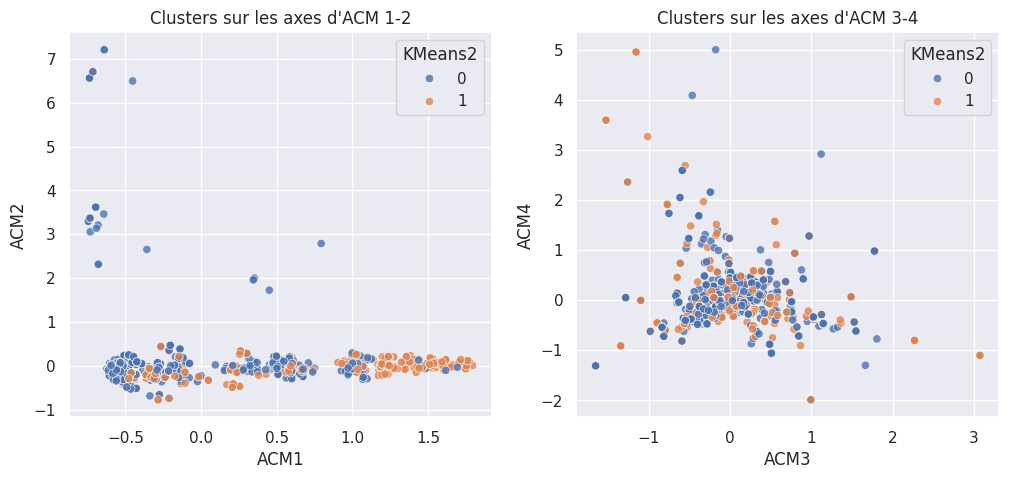
1def affiche_clusters_acm(nom_cluster):
2 _, ax = plt.subplots(1, 2, figsize=(12, 5))
3
4 ax[0].set_title("Clusters sur les axes d'ACM 1-2")
5 ax[1].set_title("Clusters sur les axes d'ACM 3-4")
6
7 sns.scatterplot(
8 composantes_acm,
9 x="ACM1",
10 y="ACM2",
11 hue=df_avec_clusters[nom_cluster],
12 alpha=0.8,
13 ax=ax[0],
14 )
15
16 sns.scatterplot(
17 composantes_acm,
18 x="ACM3",
19 y="ACM4",
20 hue=df_avec_clusters[nom_cluster],
21 alpha=0.8,
22 ax=ax[1],
23 )
24
25 plt.show()
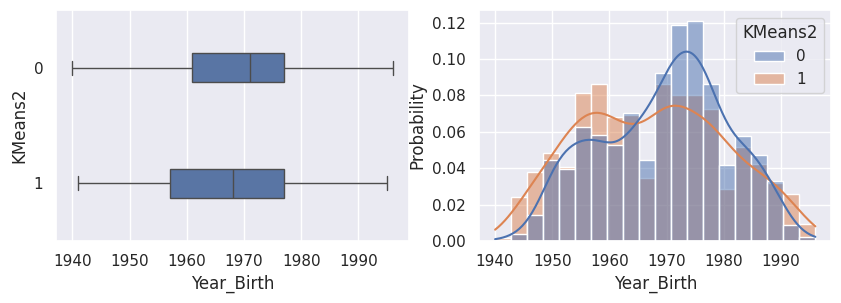
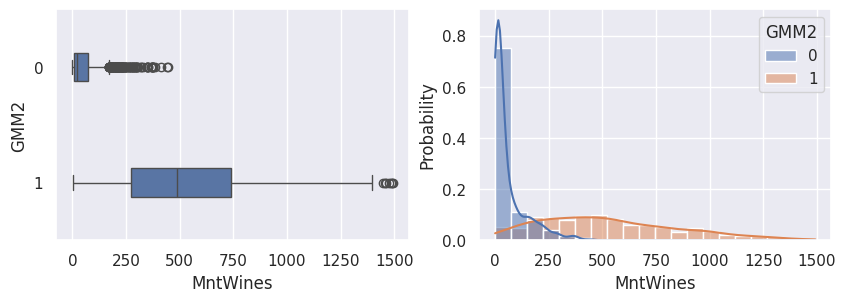
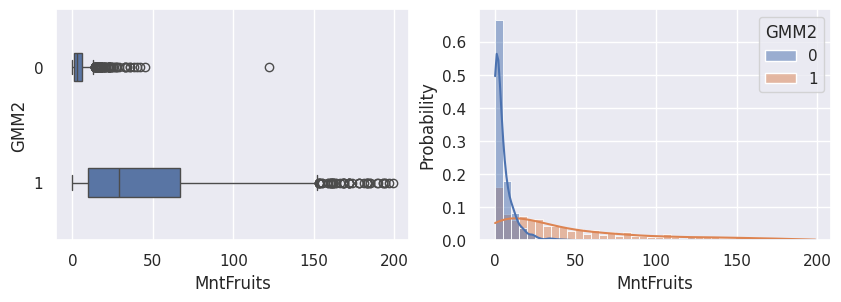
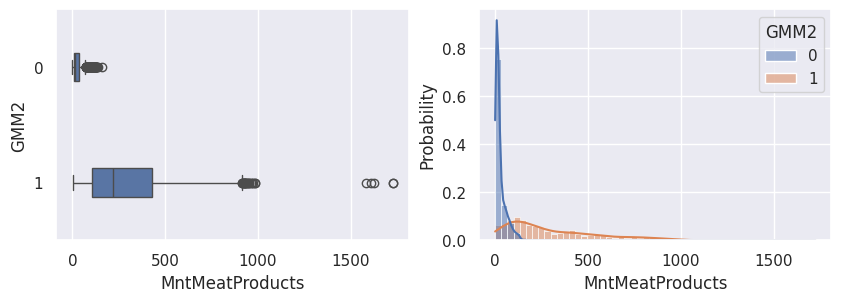
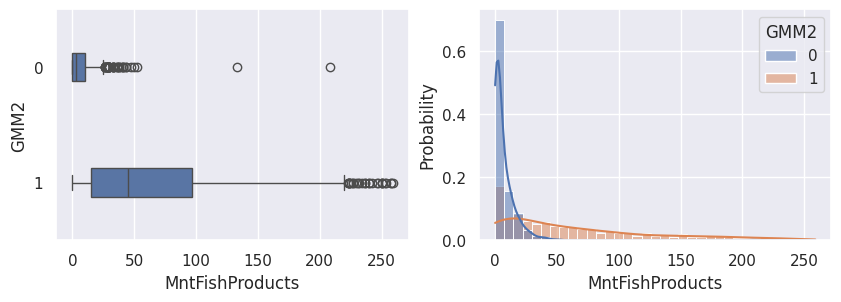
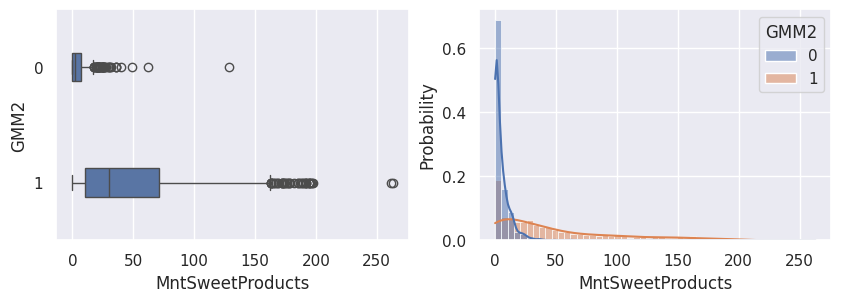
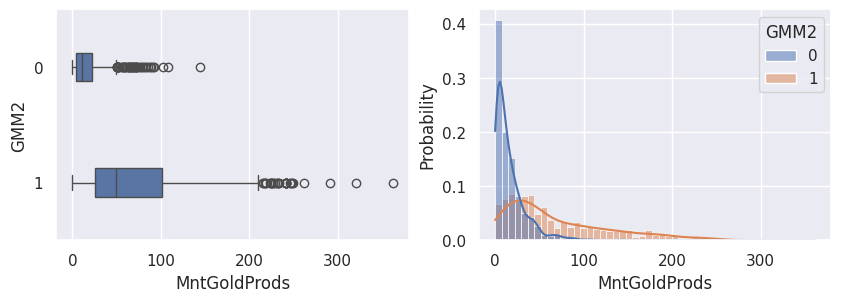
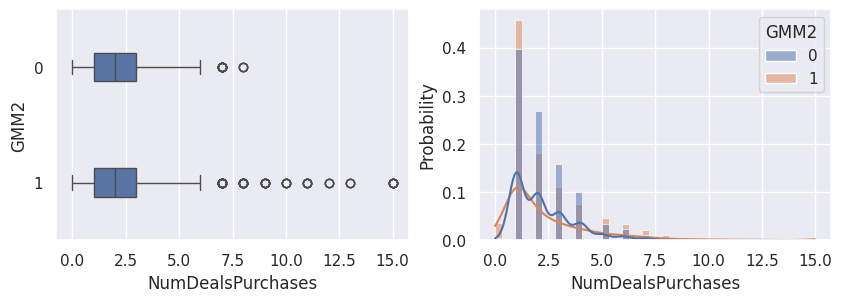
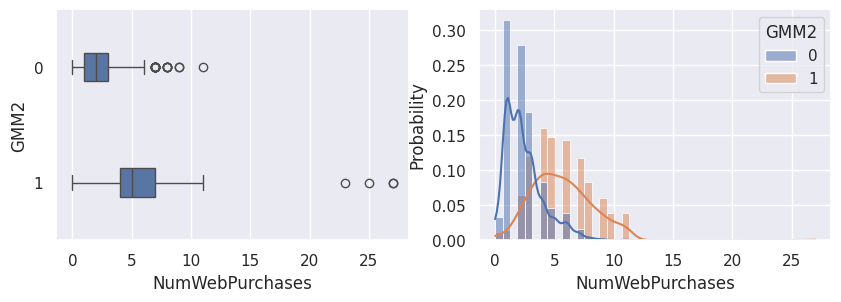
1def affiche_clusters_var_quanti(nom_cluster):
2 """Affiche les variables quantitatives en fonction des clusters."""
3 for var in var_numeriques:
4 _, ax = plt.subplots(1, 2, figsize=(10, 3))
5
6 sns.boxplot(
7 x=df[var],
8 y=df_avec_clusters[nom_cluster],
9 width=0.25,
10 ax=ax[0],
11 )
12
13 sns.histplot(
14 x=df[var],
15 kde=True,
16 ax=ax[1],
17 hue=df_avec_clusters[nom_cluster],
18 stat="probability",
19 common_norm=False,
20 )
21
22 plt.show()
1def affiche_clusters_var_quali(nom_cluster):
2 """Affiche les variables qualitatives en fonction des clusters et vice-versa."""
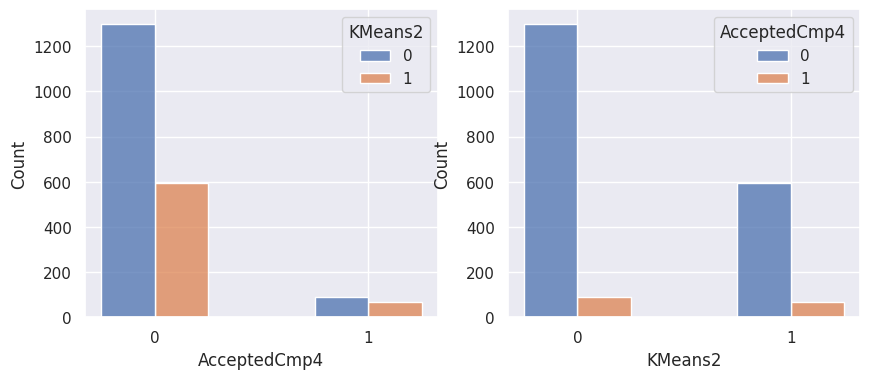
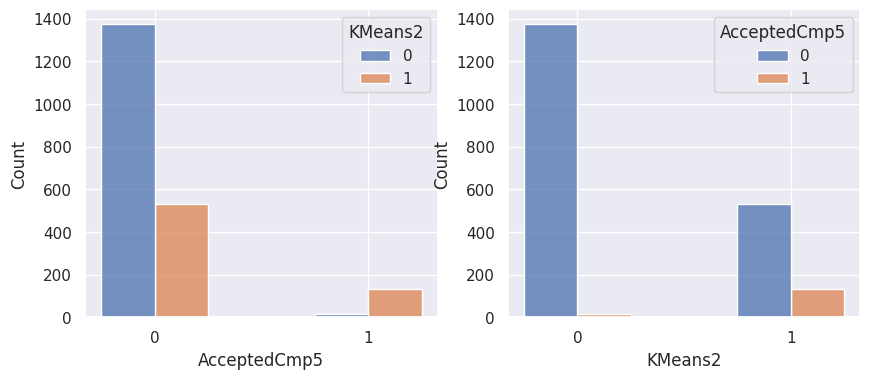
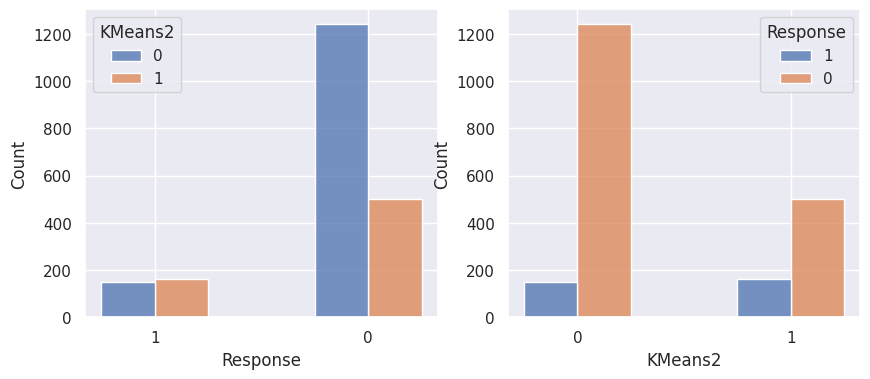
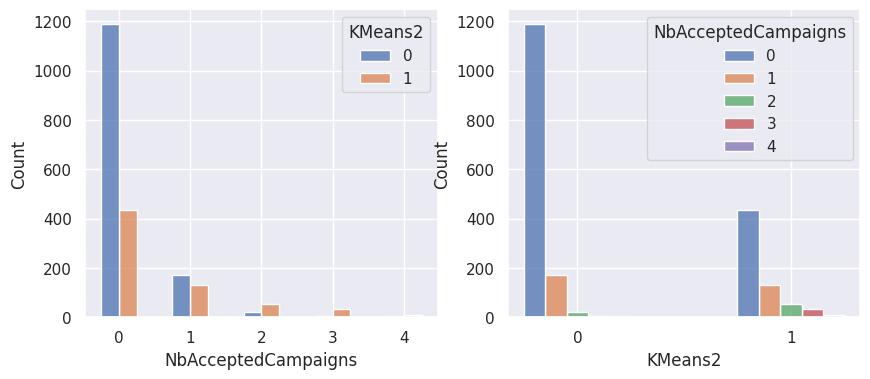
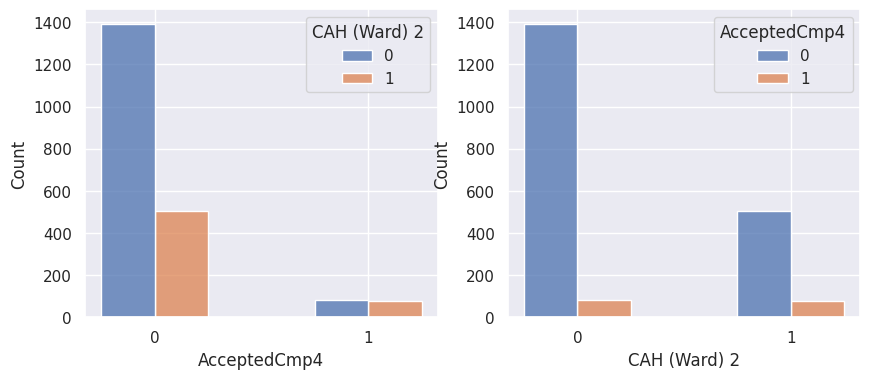
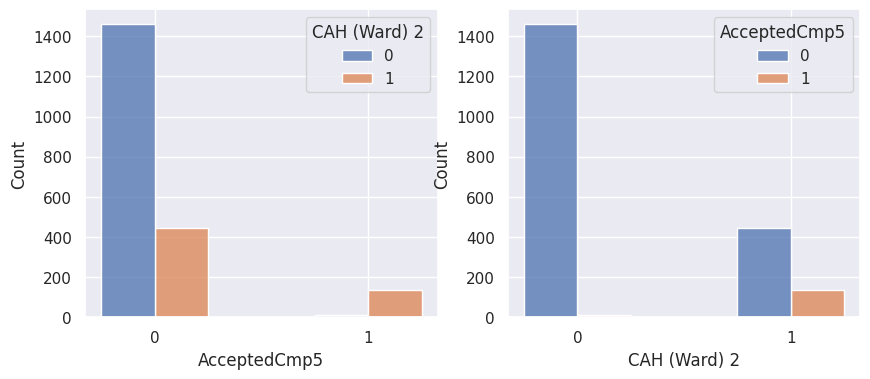
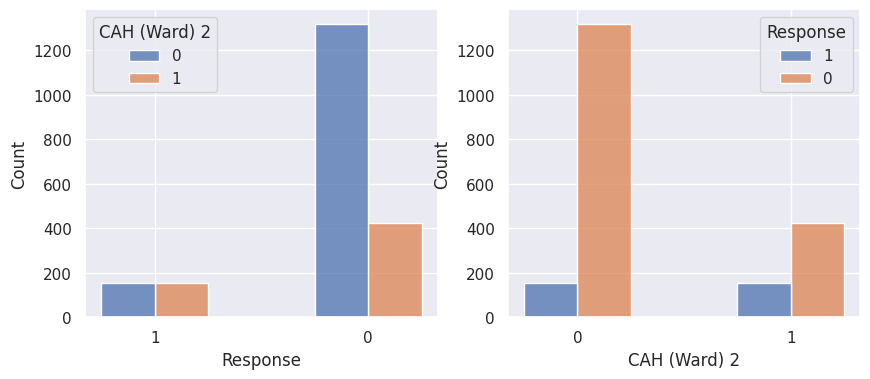
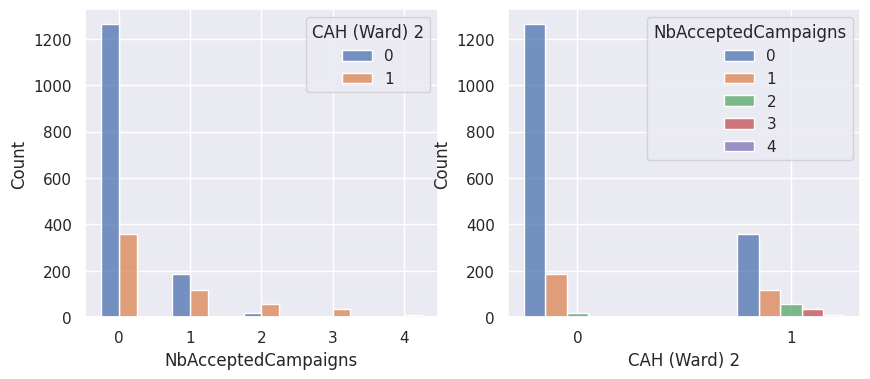
3 for var in var_categoriques_fe:
4 _, ax = plt.subplots(1, 2, figsize=(10, 4))
5
6 sns.histplot(
7 x=df[var].astype(str),
8 ax=ax[0],
9 hue=df_avec_clusters[nom_cluster],
10 multiple="dodge",
11 shrink=0.5,
12 common_norm=True,
13 )
14
15 sns.histplot(
16 hue=df[var].astype(str),
17 ax=ax[1],
18 x=df_avec_clusters[nom_cluster],
19 multiple="dodge",
20 shrink=0.5,
21 common_norm=True,
22 )
23
24 plt.show()
1def affiche_clusters(nom_cluster):
2 """Affiche les variables en fonction des clusters."""
3 affiche_taille_clusters(nom_cluster)
4 affiche_clusters_acp(nom_cluster)
5 affiche_clusters_acm(nom_cluster)
6
7 affiche_clusters_var_quanti(nom_cluster)
8 affiche_clusters_var_quali(nom_cluster)
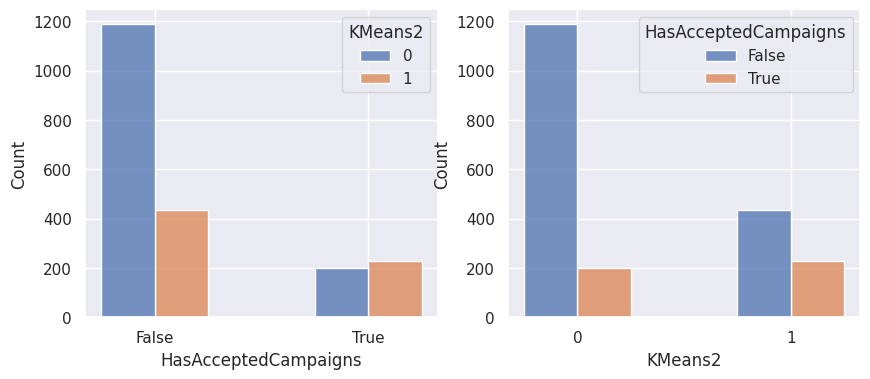
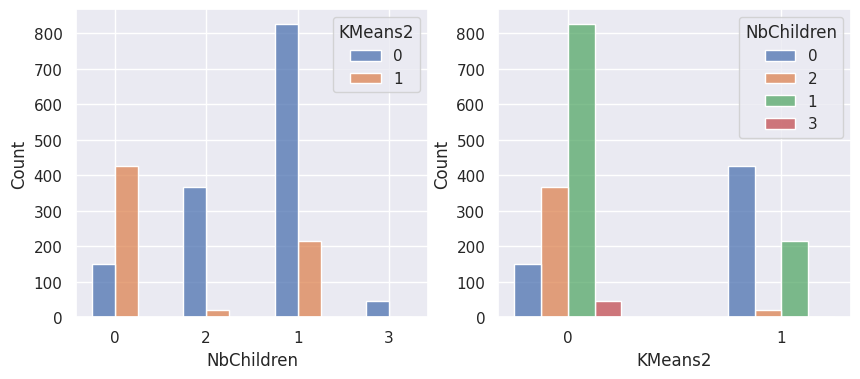
1affiche_clusters("KMeans2")
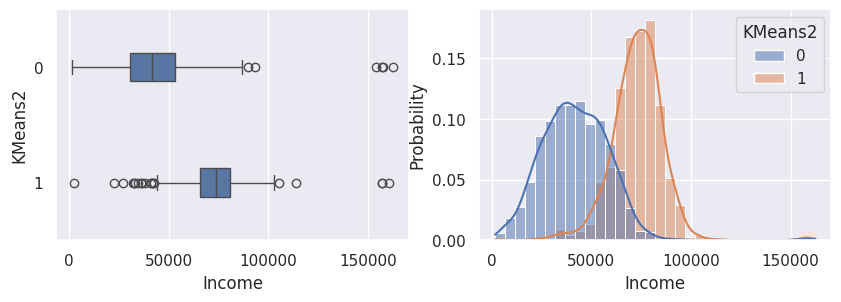
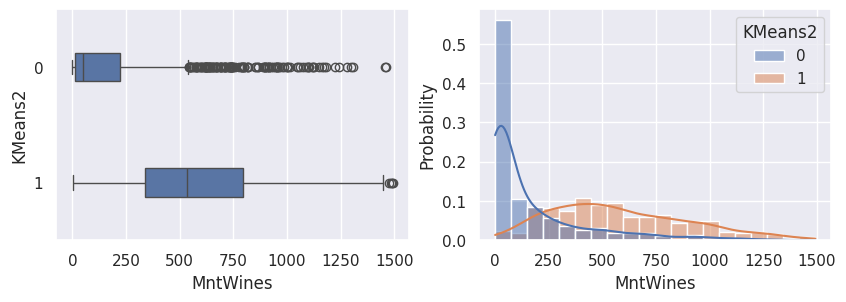
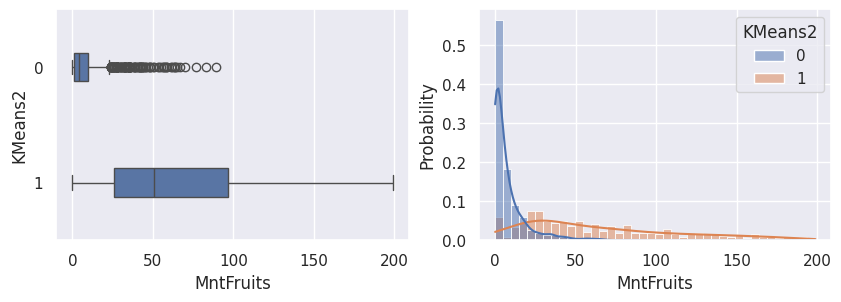
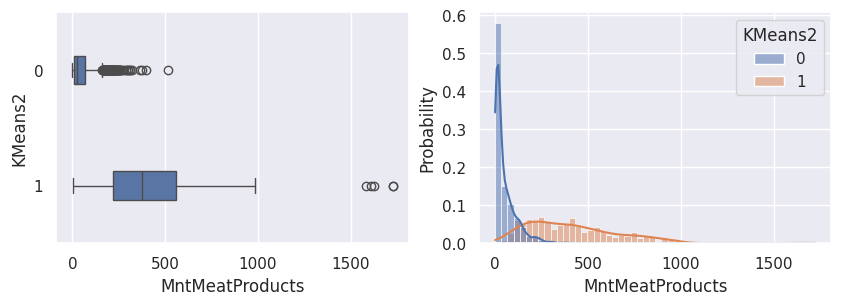
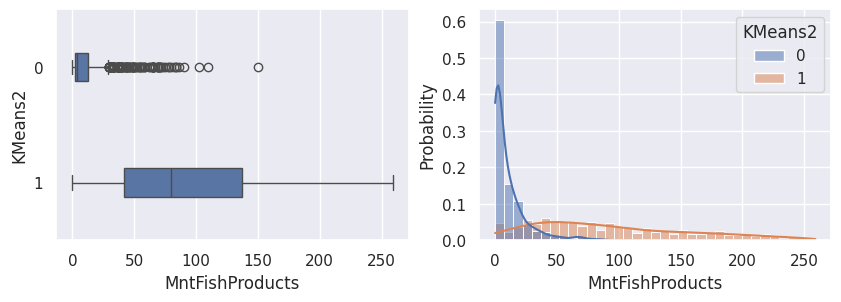
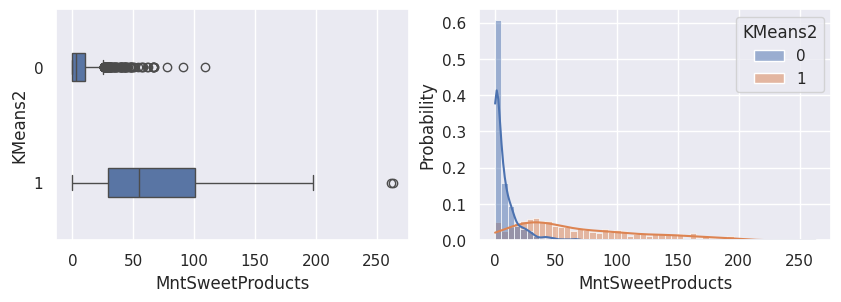
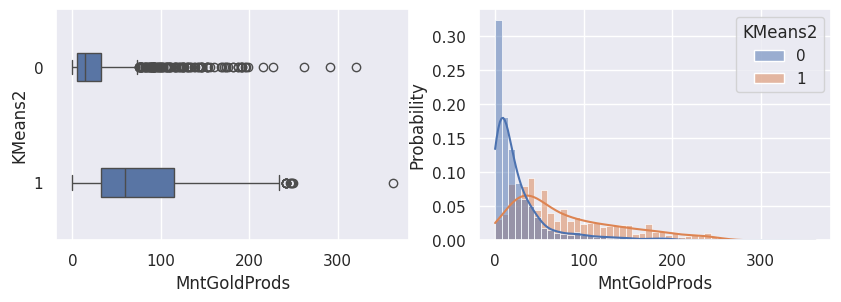
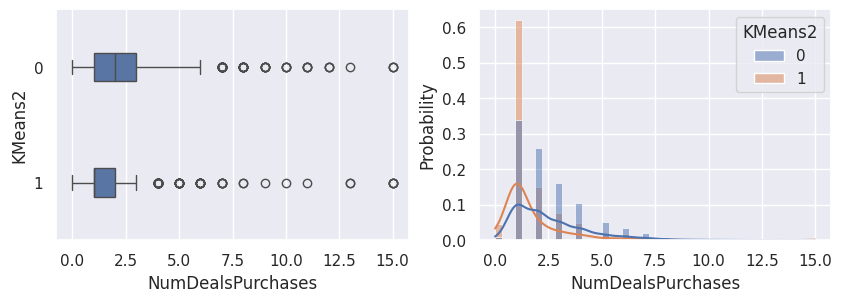
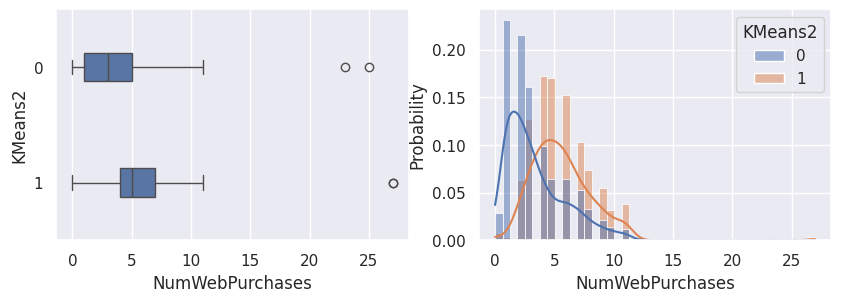
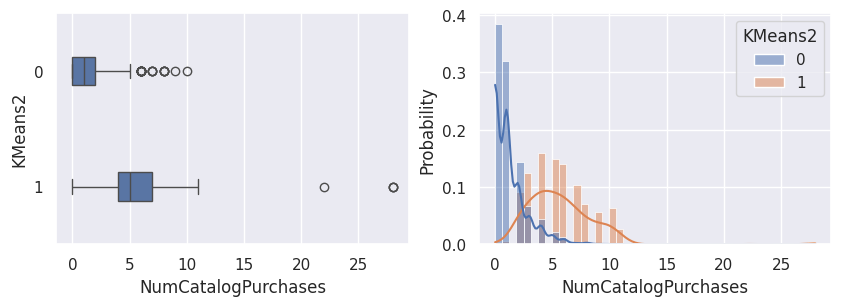
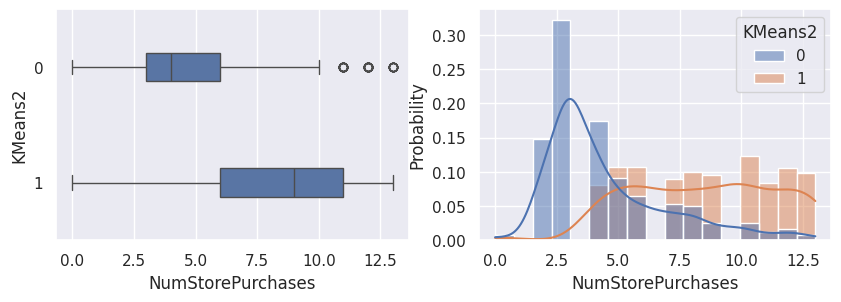
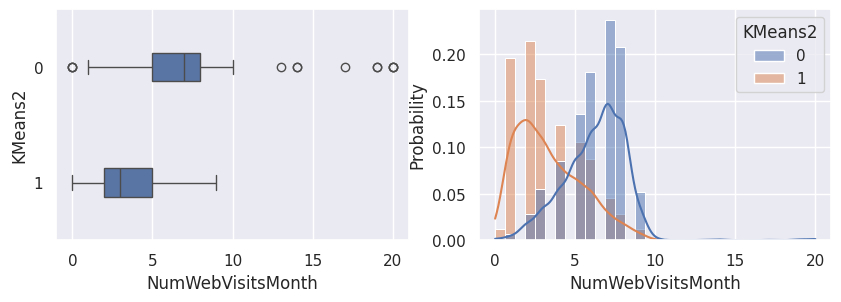
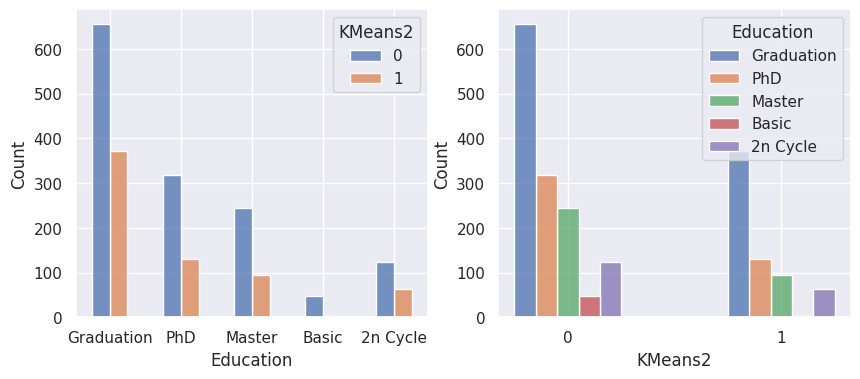
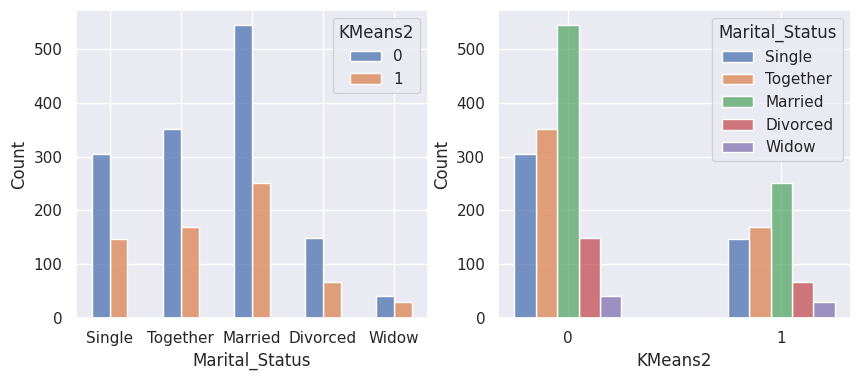
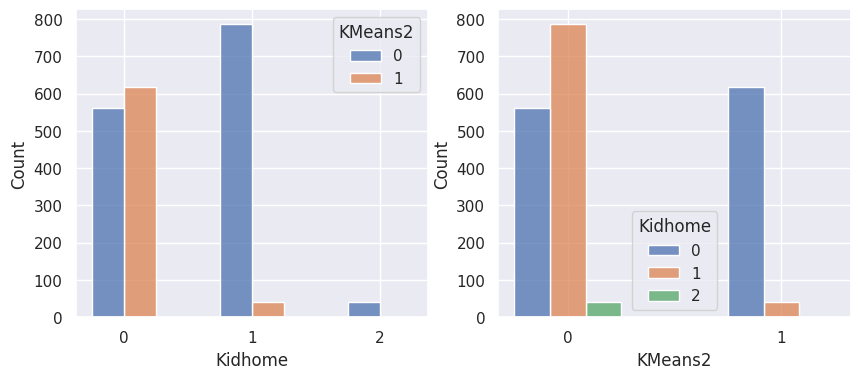
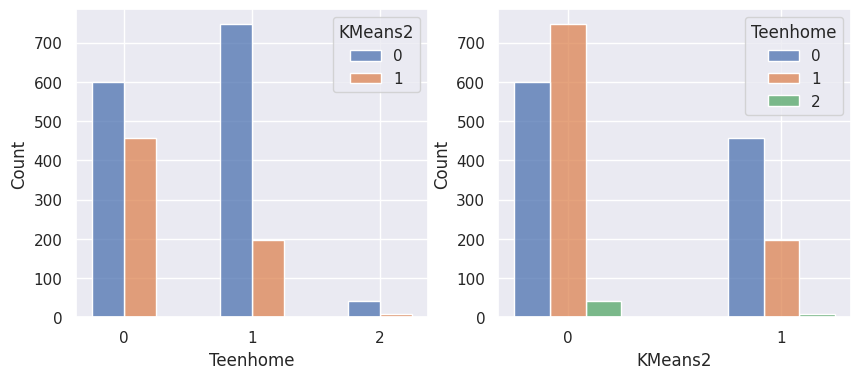
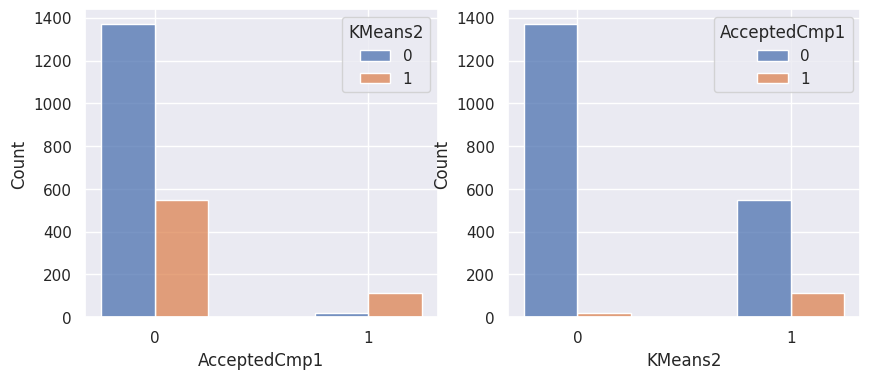
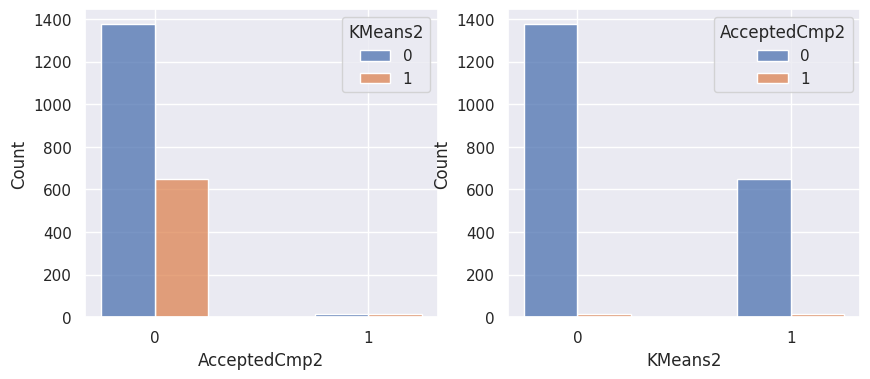
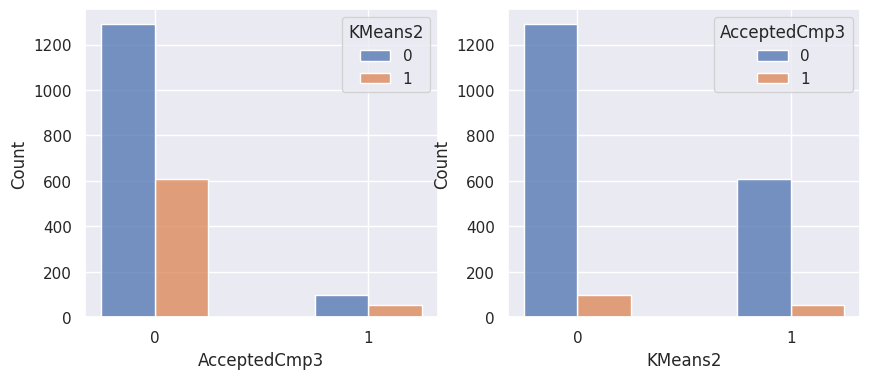
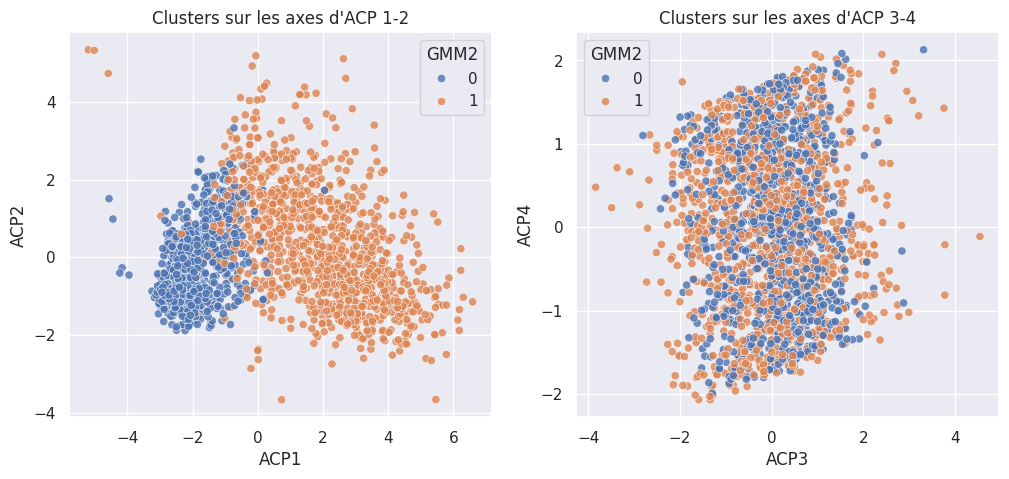
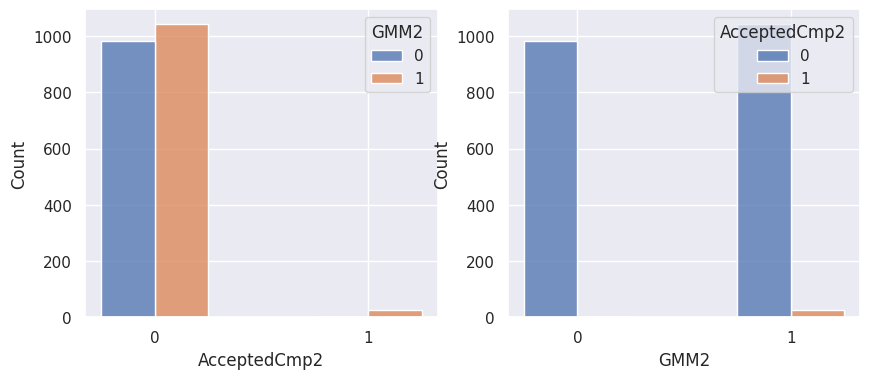
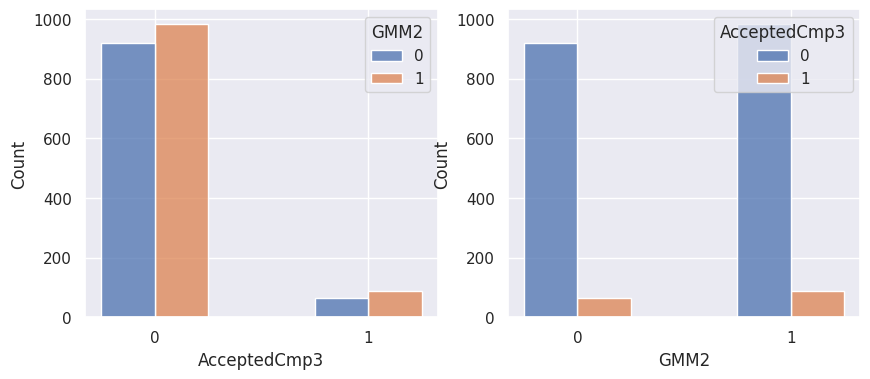
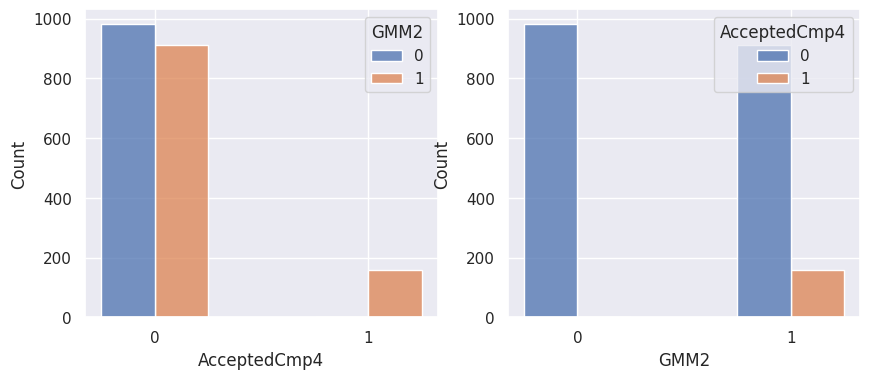
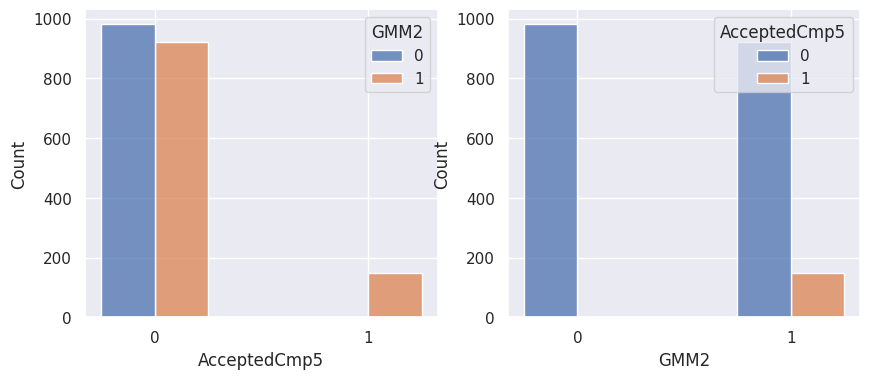
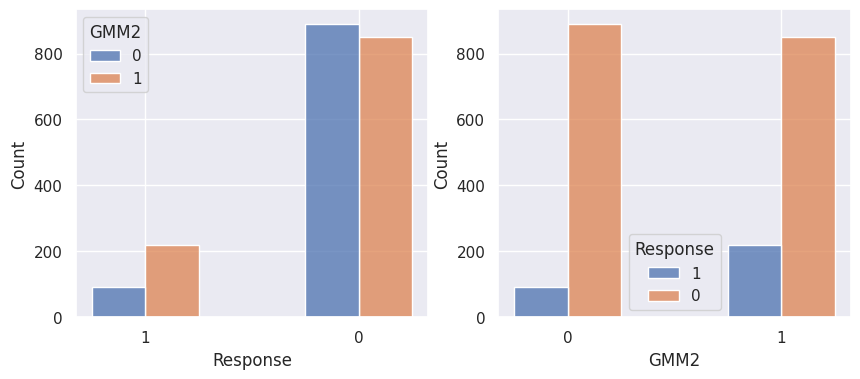
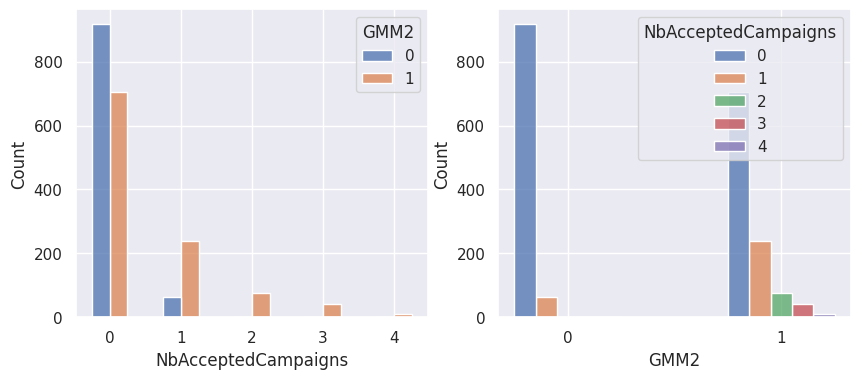
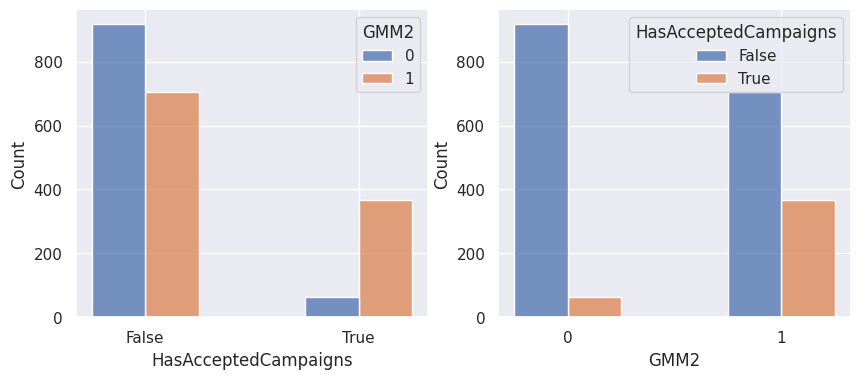
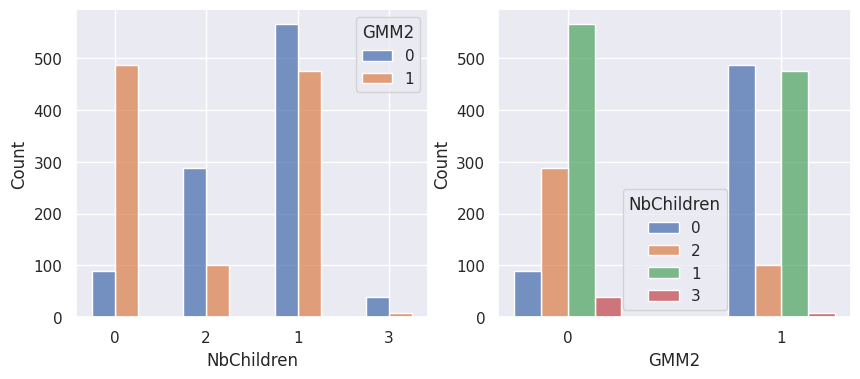
1affiche_clusters("GMM2")
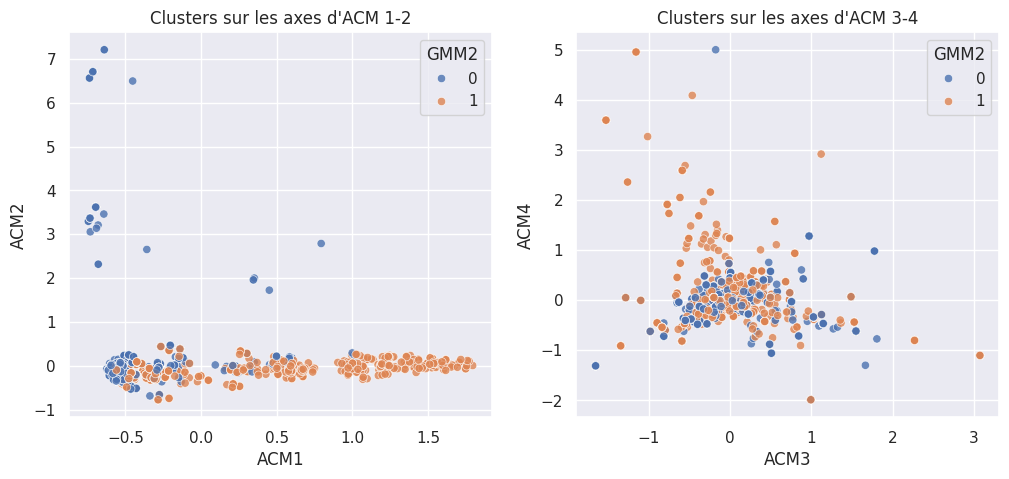
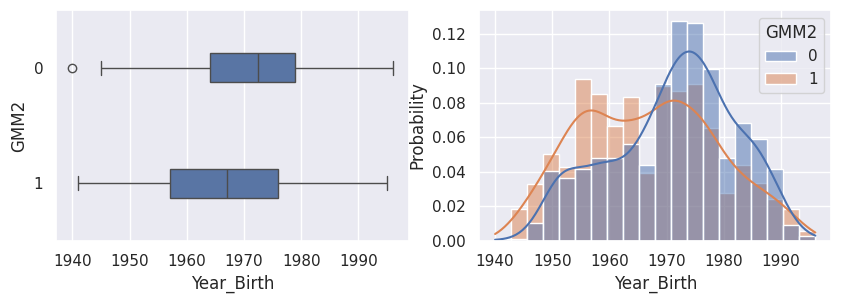
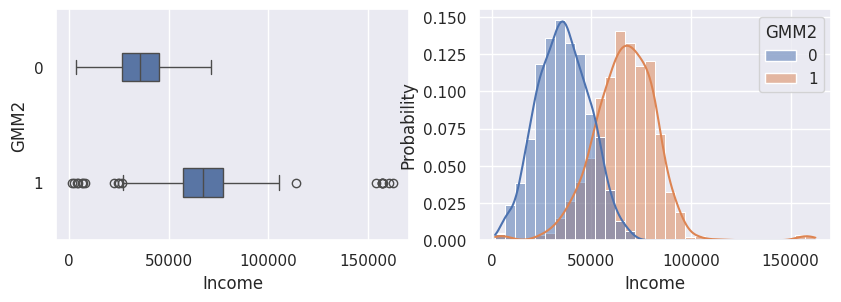
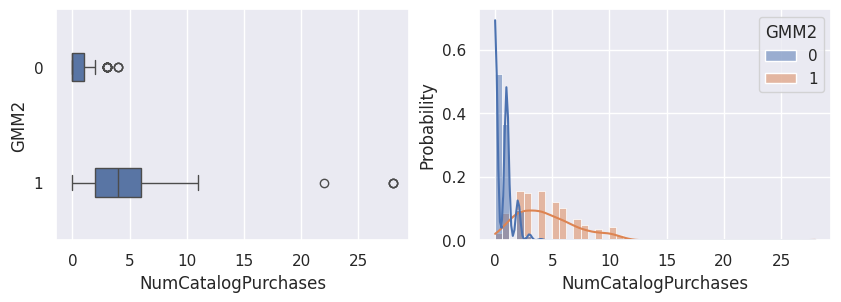
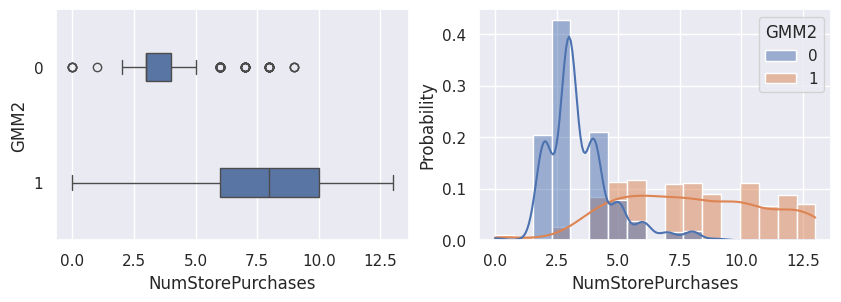
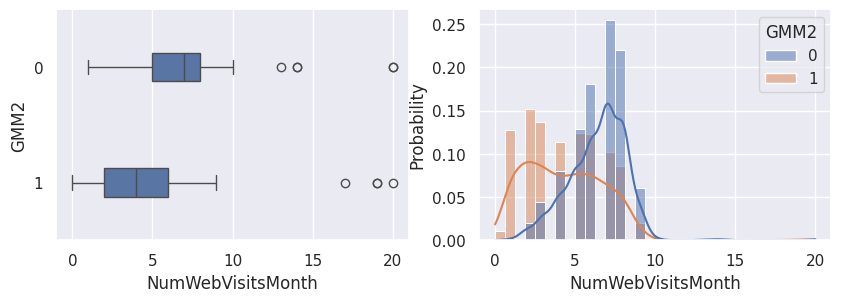
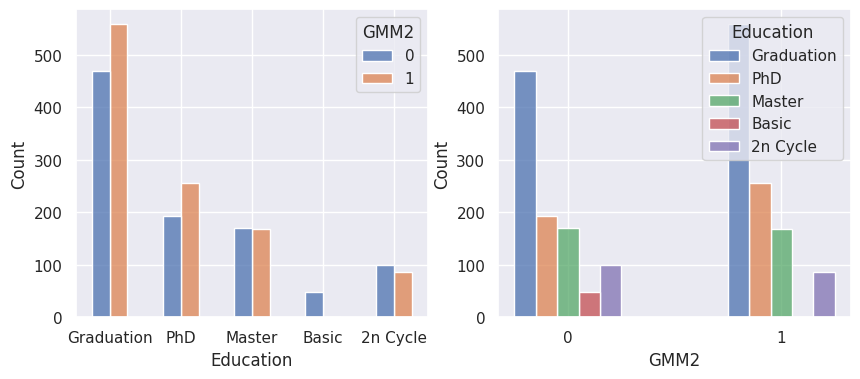
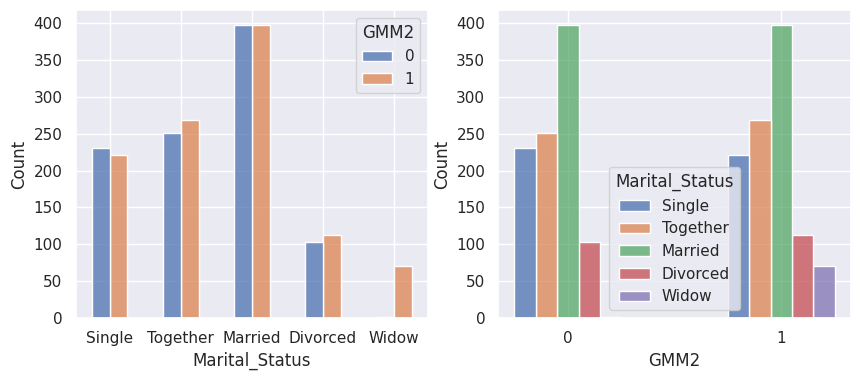
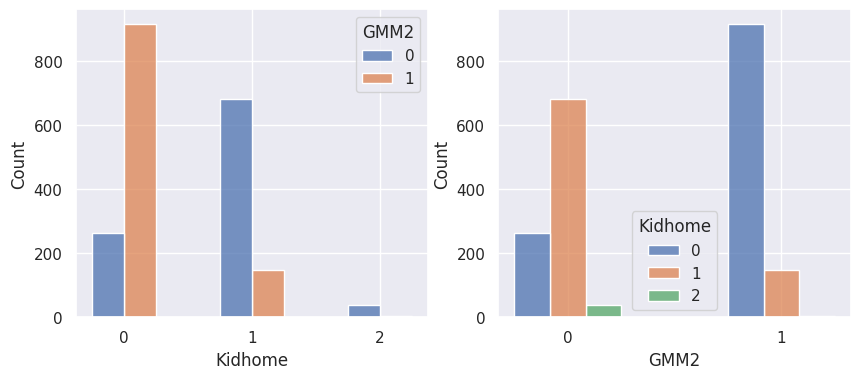
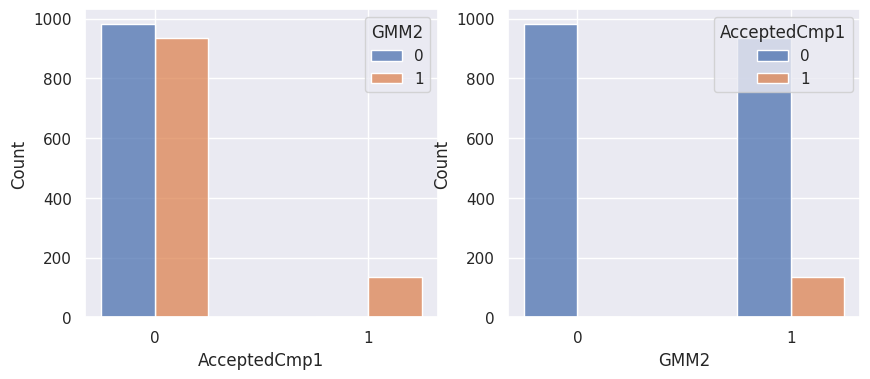
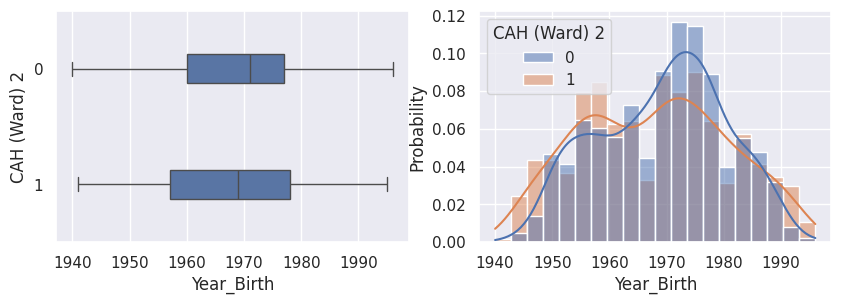
1affiche_clusters("CAH (Ward) 2")
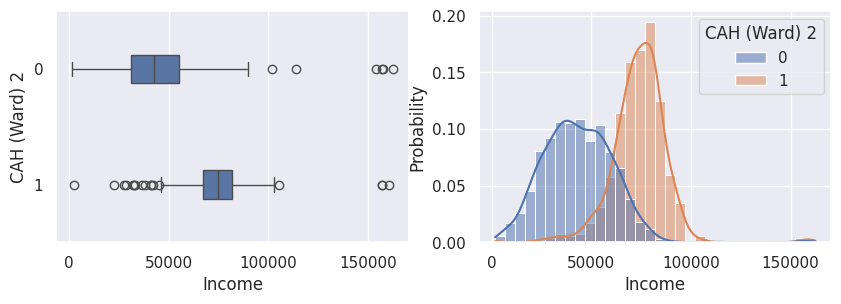
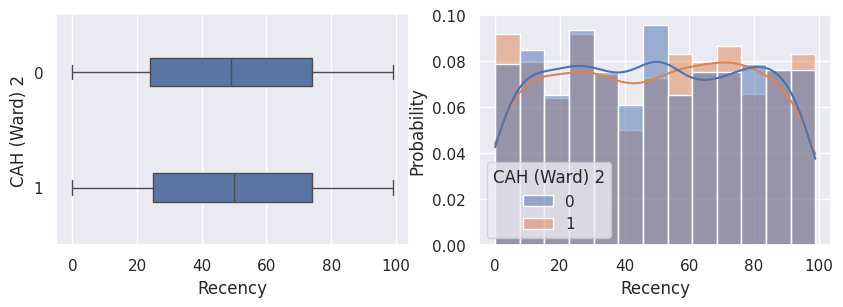
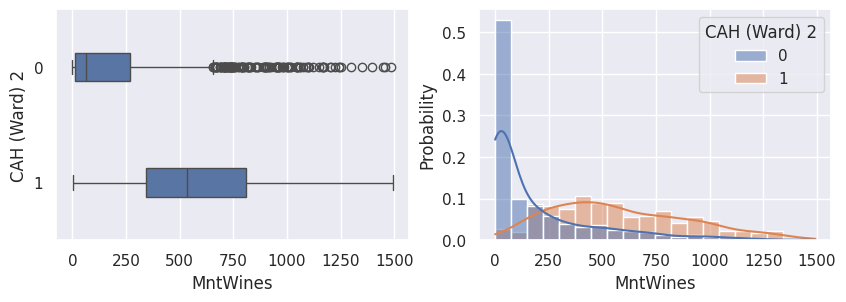
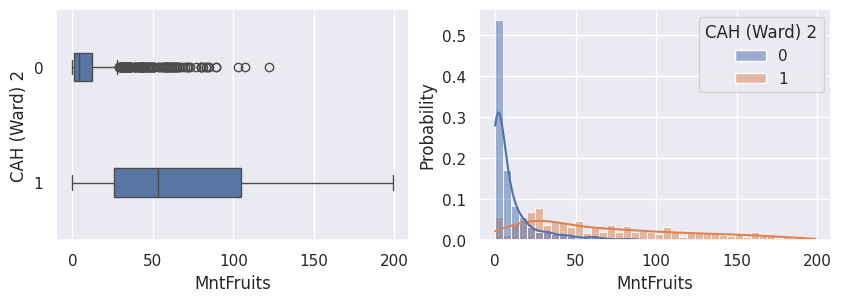
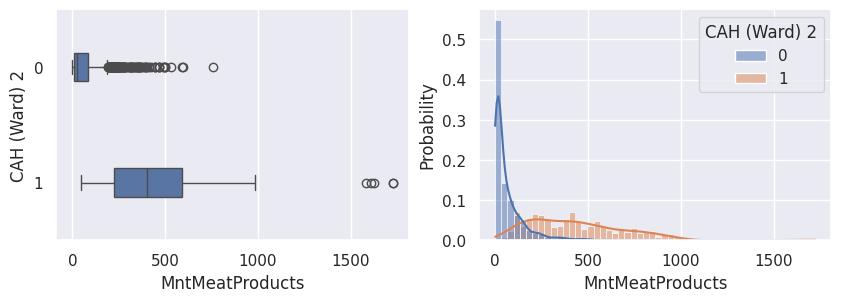
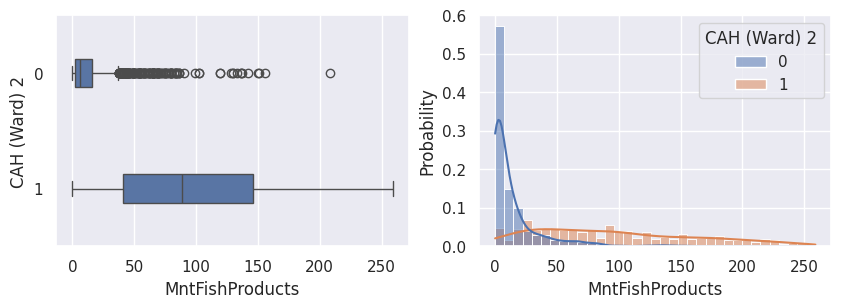
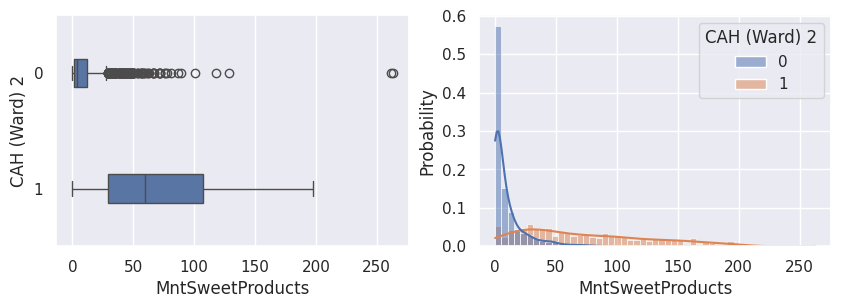
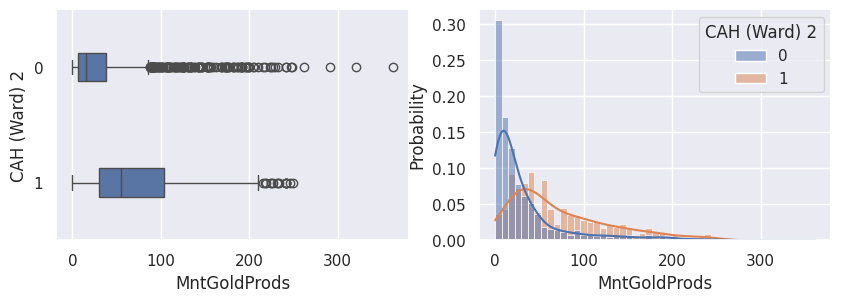
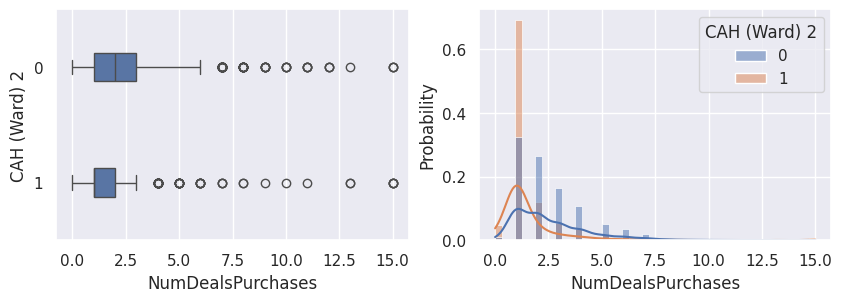
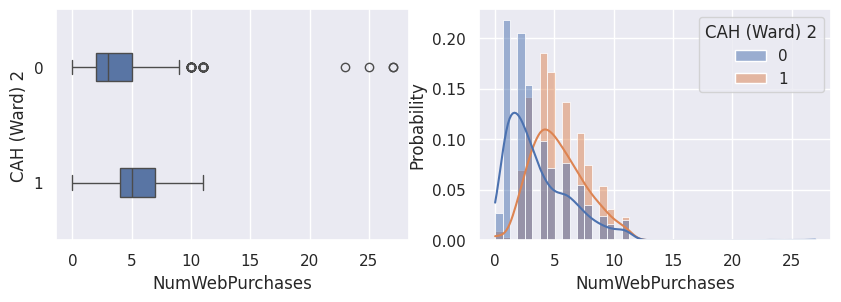
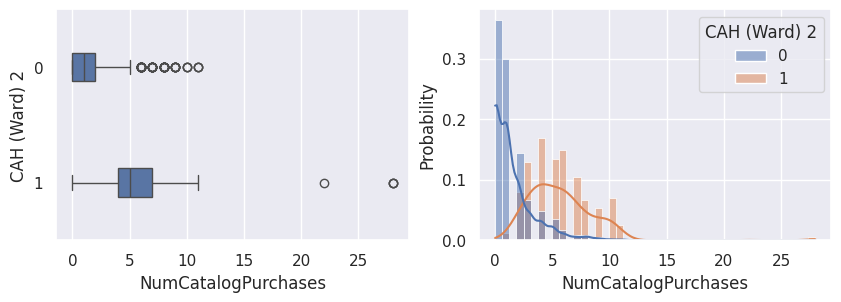
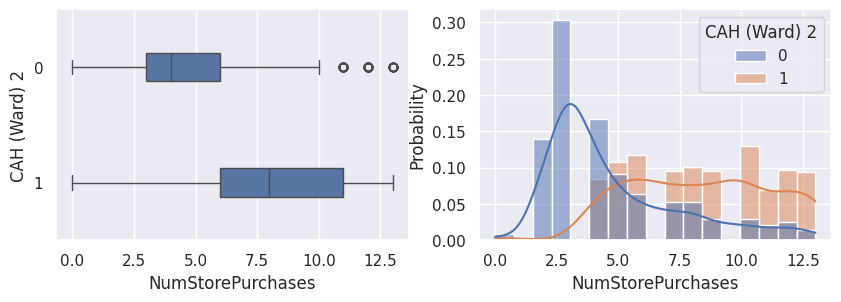
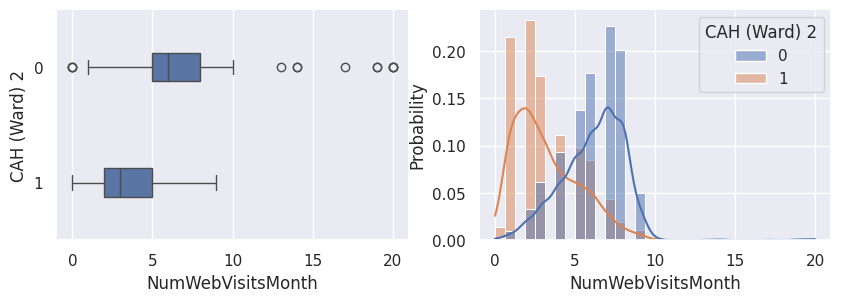
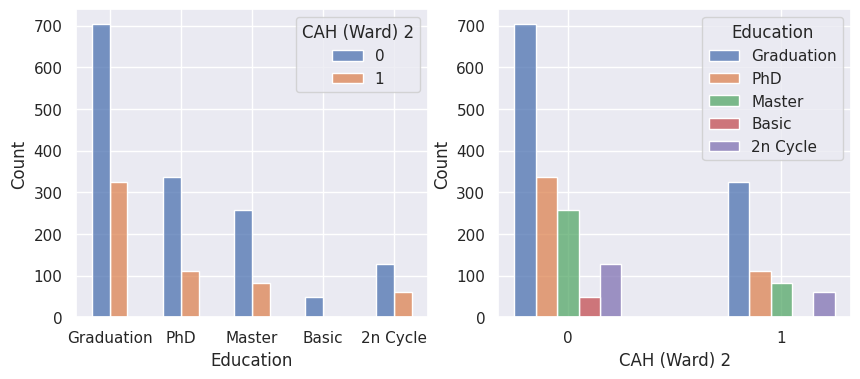
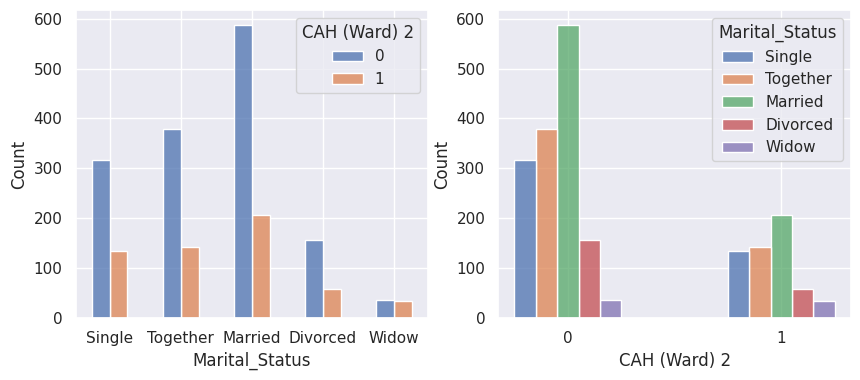
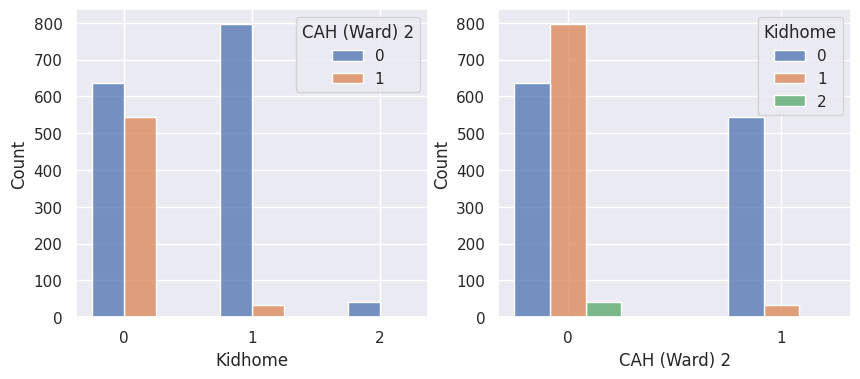
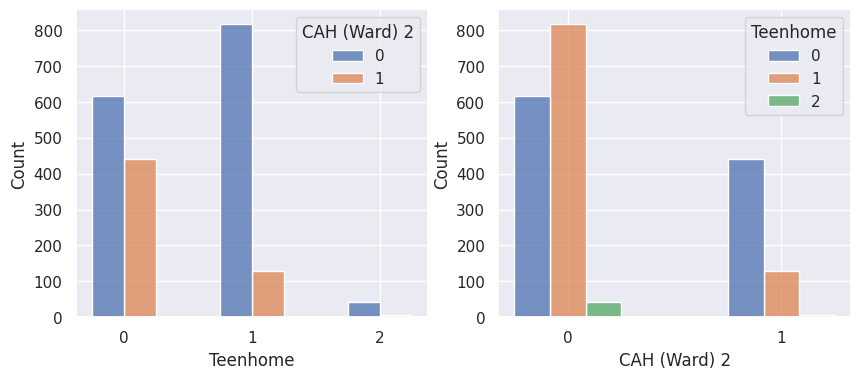
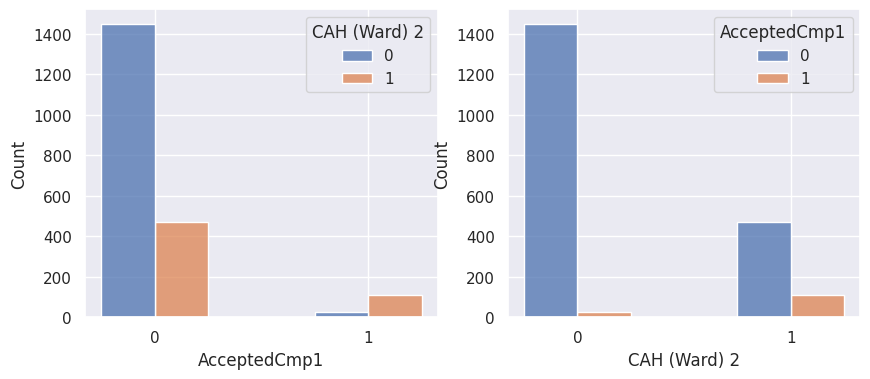
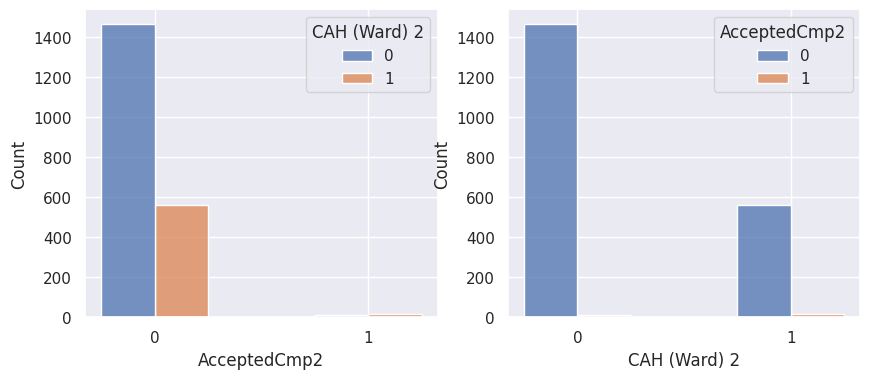
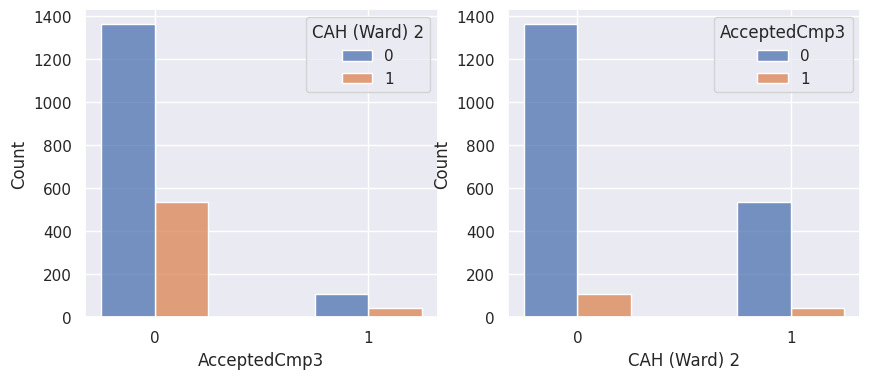
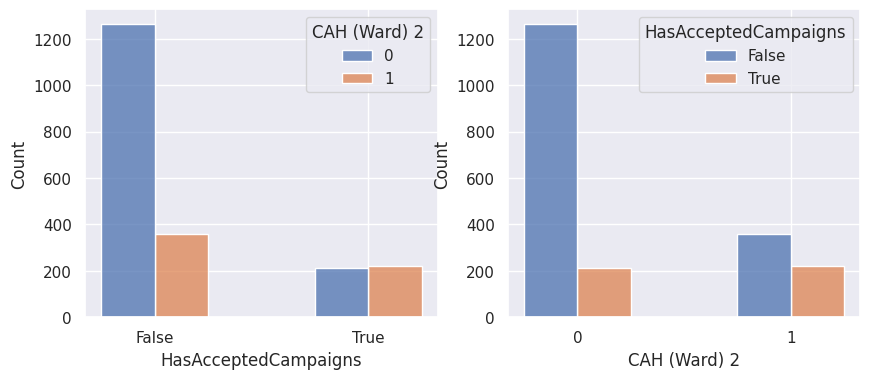
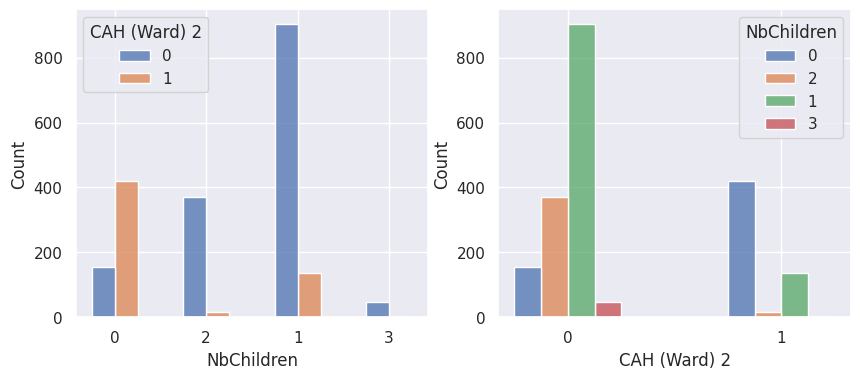
Conclusion#
Tableau. Description des clients types
Profil |
Proportion |
Education |
Revenu |
Campagnes |
Enfants |
Dépenses |
Année de |
Site Internet |
|---|---|---|---|---|---|---|---|---|
Clients qui achètent |
30% |
Bac - doctorat |
Élevé |
0-4 |
0 bas-âge |
Élevées |
1970 |
Peu de visites |
Clients qui n’achètent pas ou peu |
68% - 70% |
Brevet - doctorat |
Moyen |
0-1 |
0-3 enfants |
Peu élevées |
1970 |
Beaucoup de visites |
Clients qui n’achètent pas (n=3) |
2% |
Brevet |
Le plus faible |
0 |
0-1 bas-âge |
Aucune |
1980 |
Beaucoup de visites |
Notons aussi que parmi les clients qui achètent, la proportion d’acceptation des campagnes est beaucoup plus élevée.
Pour aller plus loin#
tester la stabilité des clusters (ici, l’initialisation des algorithmes a un impact significatif sur les clusters trouvés)
tester les différents paramètres de chacun des algorithmes de clusters pour comparaison
tester les algorithmes de clustering sur différents sous-ensembles de variables pour exhiber différents groupes
Sauvegarde des données#
1## todo: sauvegarder les clusters